from
Unifying community-wide whole-brain imaging datasets enables robust automated neuron identification and reveals determinants of neuron positioning in C. elegans. bioRxiv. 2024. Sprague DY, Rusch KW, Dunn RL, Borchardt JM, Bubnis G, Chiu GC, Wen C, Suzuki R, Chaudhary S, Dichter B, Ly R, Onami S, Lu H, Kimura K, Yemini E, Kato S.
Sleep is required to consolidate odor memory and remodel olfactory synapses. Cell. 2023 06 22; 186(13):2911-2928.e20. Chandra R, Farah F, Muñoz-Lobato F, Bokka A, Benedetti KL, Brueggemann C, Saifuddin MFA, Miller JM, Li J, Chang E, Varshney A, Jimenez V, Baradwaj A, Nassif C, Alladin S, Andersen K, Garcia AJ, Bi V, Nordquist SK, Dunn RL, Garcia V, Tokalenko K, Soohoo E, Briseno F, Kaur S, Harris M, Guillen H, Byrd D, Fung B, Bykov AE, Odisho E, Tsujimoto B, Tran A, Duong A, Daigle KC, Paisner R, Zuazo CE, Lin C, Asundi A, Churgin MA, Fang-Yen C, Bremer M, Kato S, VanHoven MK, L'Étoile ND. PMID: 37269832; PMCID: PMC10354834.
Neuroimmune cortical organoids overexpressing C4A exhibit multiple schizophrenia endophenotypes. bioRxiv. 2023. Stanton M, Modan S, Taylor P, Hariani H, Sorokin J, Rash B, Rao S, Lopez-Tobon A, Enriquez L, Dang B, Owango D, O'Neill S, Castrillo C, Nicola J, Ye K, Blattner, R, Gonzalez F, Antonio D, Ramkumar P, Lash A, Flanzer D, Bardehle S, Gyoneva S, Shah K, Kato S, Skibinski G.
Implicitization of biquadratic Bezier triangle and quadrilateral surfaces. arXiv. 2023. Borchardt J and Kato S.
A set of hub neurons and non-local connectivity features support global brain dynamics in C. elegans. Curr Biol. 2022 08 22; 32(16):3443-3459.e8. Uzel K, Kato S, Zimmer M. PMID: 35809568.
Hierarchical confounder discovery in the experiment-machine learning cycle. Cell Patterns. 2022. Rogozhnikov A, Ramkumar P, Bedi R, Kato S, Escola GS.
Information theory rules out the reflex-chain model of C. elegans locomotion. bioRxiv. 2022. Webb J and Kato S.
Demuxalot: scaled up genetic demultiplexing for single-cell sequencing. bioRxiv. 2021. Rogozhnikov A, Ramkumar P, Shah K, Bedi R, Kato S, Escola GS.
minimo: a linked data and metadata storage system for small labs. Journal of Open Source Software. 2021; 6(60):2979. Borchardt J, Dunn R, and Kato S.
Pycro-Manager: open-source software for customized and reproducible microscope control. Nat Methods. 2021 03; 18(3):226-228. Pinkard H, Stuurman N, Ivanov IE, Anthony NM, Ouyang W, Li B, Yang B, Tsuchida MA, Chhun B, Zhang G, Mei R, Anderson M, Shepherd DP, Hunt-Isaak I, Dunn RL, Jahr W, Kato S, Royer LA, Thiagarajah JR, Eliceiri KW, Lundberg E, Mehta SB, Waller L. PMID: 33674797; PMCID: PMC8532176.
Optimization and scaling of patient-derived brain organoids uncovers deep phenotypes of disease. bioRxiv. 2020. Shah K, Bedi R, Ramkumar P, Tong Z, Rash B, Stanton B, Sorokin J, Apaydin C, Batarse A, Bergamaschi J, Blattner R, Brown S, Bosshardt A, Castrillo C, Dang B, Drusinksy S, Enriquez L, Grayson D, Hilliard J, Hsu P-K, Johnson C, Jones R, Lash A, Lee C-Y, Li K, McKay A, Mount E, Nicola J, Oumzil I, Paek J, Pascoe D, Piepho A, Poust S, Quang D, Schultz M, Sims J, Taylor P, Treiman G, Wueseke O, Young N, Pollen A, Flanzer D, Chao D, Skibinski G, Kato S, Escola GS.
Soma-Targeted Imaging of Neural Circuits by Ribosome Tethering. Neuron. 2020 08 05; 107(3):454-469.e6. Chen Y, Jang H, Spratt PWE, Kosar S, Taylor DE, Essner RA, Bai L, Leib DE, Kuo TW, Lin YC, Patel M, Subkhangulova A, Kato S, Feinberg EH, Bender KJ, Knight ZA, Garrison JL. PMID: 32574560; PMCID: PMC7540731.
A probabilistic atlas for cell identification. arXiv. 2019. Bubnis G, Ban S, Difranco MD, Kato S.
Regulation of two motor patterns enables the gradual adjustment of locomotion strategy in Caenorhabditis elegans. Elife. 2016 05 25; 5. Hums I, Riedl J, Mende F, Kato S, Kaplan HS, Latham R, Sonntag M, Traunmuller L, Zimmer M. PMID: 27222228; PMCID: PMC4880447.
Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell. 2015 Oct 22; 163(3):656-69. Kato S, Kaplan HS, Schrödel T, Skora S, Lindsay TH, Yemini E, Lockery S, Zimmer M. PMID: 26478179.
Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat Methods. 2014 Jul; 11(7):727-730. Prevedel R, Yoon YG, Hoffmann M, Pak N, Wetzstein G, Kato S, Schrödel T, Raskar R, Zimmer M, Boyden ES, Vaziri A. PMID: 24836920; PMCID: PMC4100252.
Temporal responses of C. elegans chemosensory neurons are preserved in behavioral dynamics. Neuron. 2014 Feb 05; 81(3):616-28. Kato S, Xu Y, Cho CE, Abbott LF, Bargmann CI. PMID: 24440227; PMCID: PMC4112952.
Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. 2010 May; 13(5):615-21. Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, Bargmann CI. PMID: 20364145; PMCID: PMC2937567.
Soma-targeted imaging of neural circuits by ribosome tethering
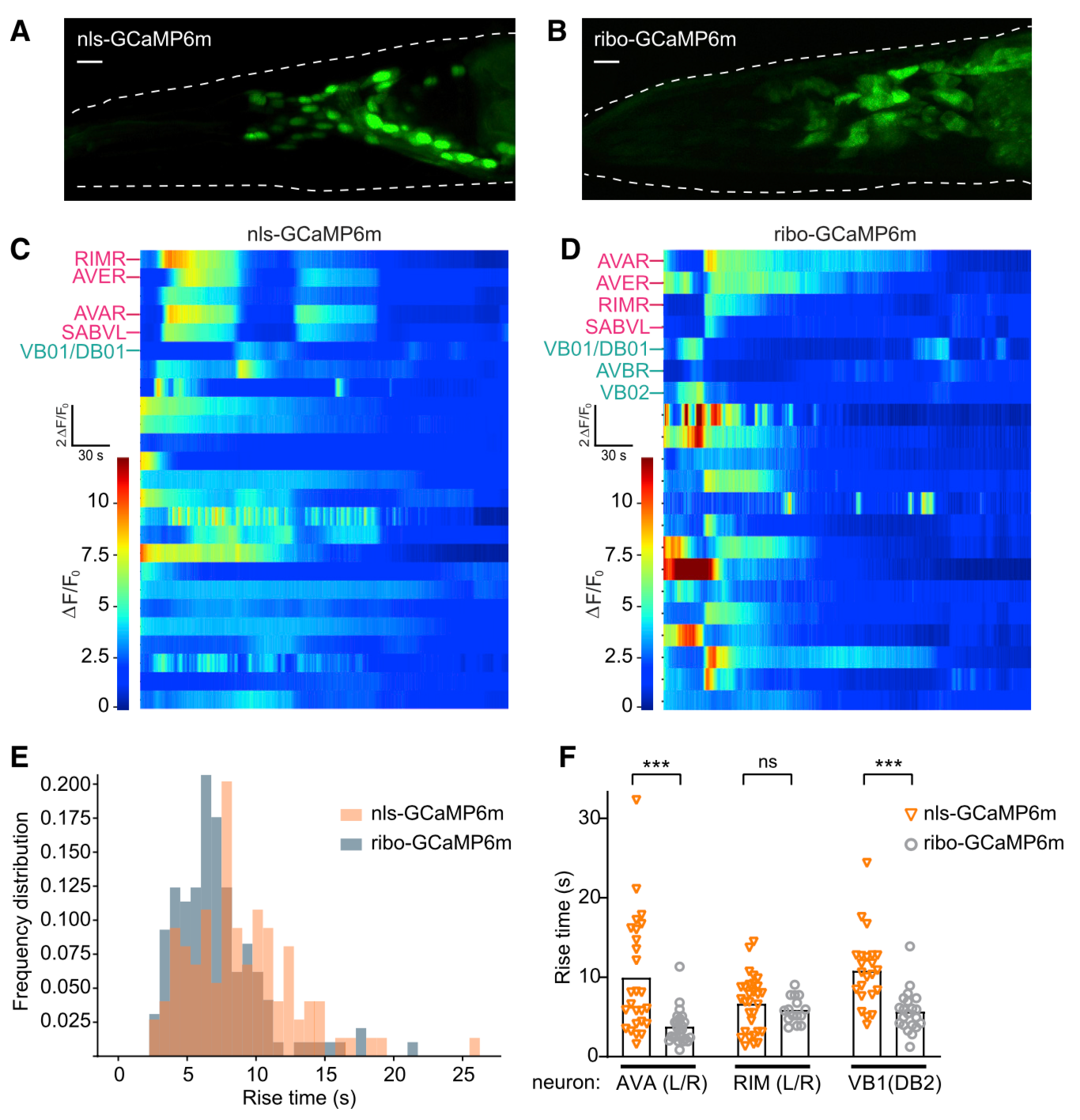 Neuroscience relies on techniques for imaging the structure and dynamics of neural circuits, but the cell bodies of individual neurons are often obscured by overlapping fluorescence from axons and dendrites in surrounding neuropil. Here, we describe two strategies for using the ribosome to restrict the expression of fluorescent proteins to the neuronal soma. We show first that a ribosome-tethered nanobody can be used to trap GFP in the cell body, thereby enabling direct visualization of previously undetectable GFP fluorescence. We then design a ribosome-tethered GCaMP for imaging calcium dynamics. We show that this reporter faithfully tracks somatic calcium dynamics in the mouse brain while eliminating cross-talk between neurons caused by contaminating neuropil. In worms, this reporter enables whole-brain imaging with faster kinetics and brighter fluorescence than commonly used nuclear GCaMPs. These two approaches provide a general way to enhance the specificity of imaging in neurobiology.
Neuroscience relies on techniques for imaging the structure and dynamics of neural circuits, but the cell bodies of individual neurons are often obscured by overlapping fluorescence from axons and dendrites in surrounding neuropil. Here, we describe two strategies for using the ribosome to restrict the expression of fluorescent proteins to the neuronal soma. We show first that a ribosome-tethered nanobody can be used to trap GFP in the cell body, thereby enabling direct visualization of previously undetectable GFP fluorescence. We then design a ribosome-tethered GCaMP for imaging calcium dynamics. We show that this reporter faithfully tracks somatic calcium dynamics in the mouse brain while eliminating cross-talk between neurons caused by contaminating neuropil. In worms, this reporter enables whole-brain imaging with faster kinetics and brighter fluorescence than commonly used nuclear GCaMPs. These two approaches provide a general way to enhance the specificity of imaging in neurobiology.
A probabilistic atlas for cell identification
We propose a general framework for a collaborative machine learning system to assist bioscience researchers with the task of labeling specific cell identities from microscopic still or video imaging. The distinguishing features of this approach versus prior approaches include: (1) use of a statistical model of cell features that is iteratively improved, (2) generation of probabilistic guesses at cell ID rather than single best-guesses for each cell, (3) tracking of joint probabilities of features within and across cells, and (4) ability to exploit multi-modal features, such as cell position, morphology, reporter intensities, and activity. We provide an example implementation of such a system applicable to labeling fluorescently tagged C. elegans neurons. As a proof of concept, we use a generative spring-mass model to simulate sequences of cell imaging datasets with variable cell positions and fluorescence intensities. Training on synthetic data, we find that atlases that track inter-cell positional correlations give higher labeling accuracies than those that treat cell positions independently. Tracking an additional feature type, fluorescence intensity, boosts accuracy relative to a position-only atlas, suggesting that multiple cell features could be leveraged to improve automated label predictions.
Global brain dynamics embed the motor command sequence of Caenorhabditis elegans.
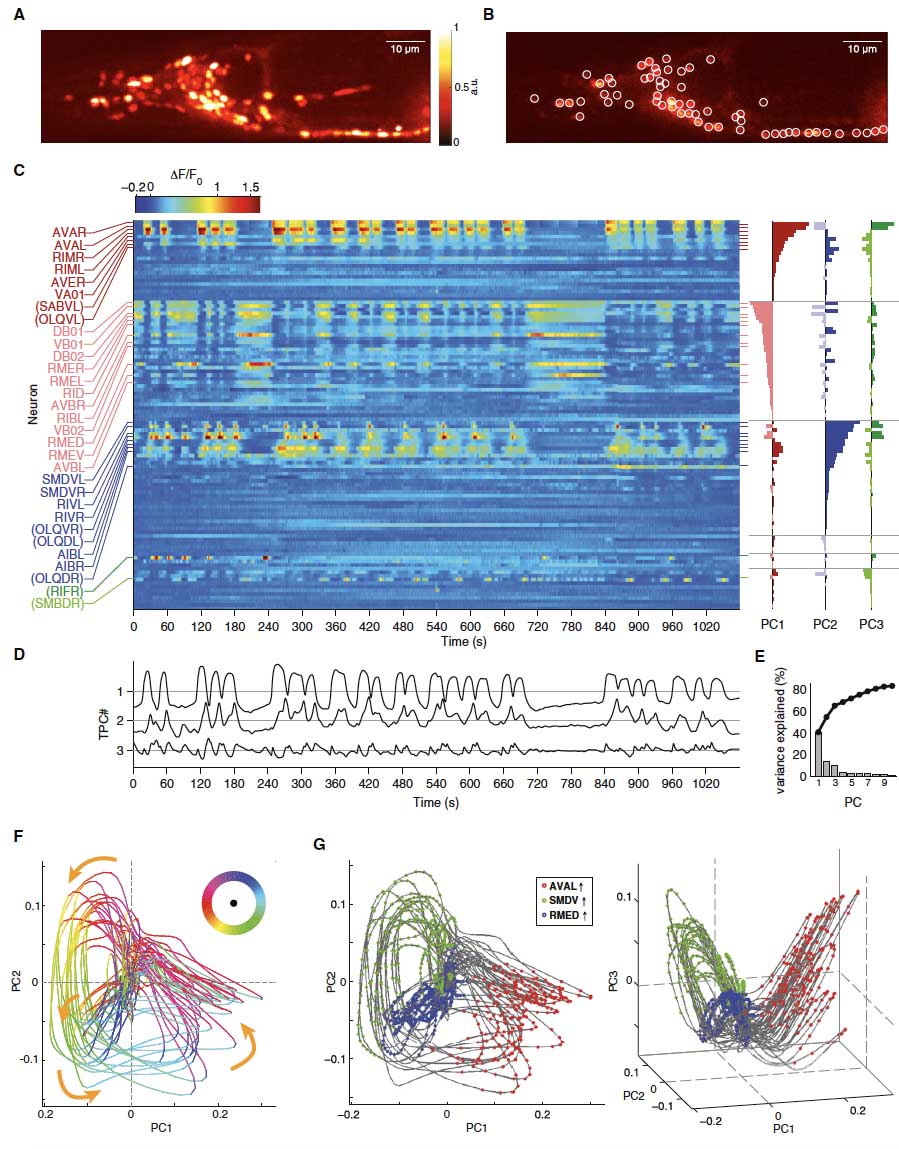 While isolated motor actions can be correlated with activities of neuronal networks, an unresolved problem is how the brain assembles these activities into organized behaviors like action sequences. Using brain-wide calcium imaging in Caenorhabditis elegans, we show that a large proportion of neurons across the brain share information by engaging in coordinated, dynamical network activity. This brain state evolves on a cycle, each segment of which recruits the activities of different neuronal sub-populations and can be explicitly mapped, on a single trial basis, to the animals' major motor commands. This organization defines the assembly of motor commands into a string of run-and-turn action sequence cycles, including decisions between alternative behaviors. These dynamics serve as a robust scaffold for action selection in response to sensory input. This study shows that the coordination of neuronal activity patterns into global brain dynamics underlies the high-level organization of behavior.
While isolated motor actions can be correlated with activities of neuronal networks, an unresolved problem is how the brain assembles these activities into organized behaviors like action sequences. Using brain-wide calcium imaging in Caenorhabditis elegans, we show that a large proportion of neurons across the brain share information by engaging in coordinated, dynamical network activity. This brain state evolves on a cycle, each segment of which recruits the activities of different neuronal sub-populations and can be explicitly mapped, on a single trial basis, to the animals' major motor commands. This organization defines the assembly of motor commands into a string of run-and-turn action sequence cycles, including decisions between alternative behaviors. These dynamics serve as a robust scaffold for action selection in response to sensory input. This study shows that the coordination of neuronal activity patterns into global brain dynamics underlies the high-level organization of behavior.
Regulation of two motor patterns enables the gradual adjustment of locomotion strategy in Caenorhabditis elegans.
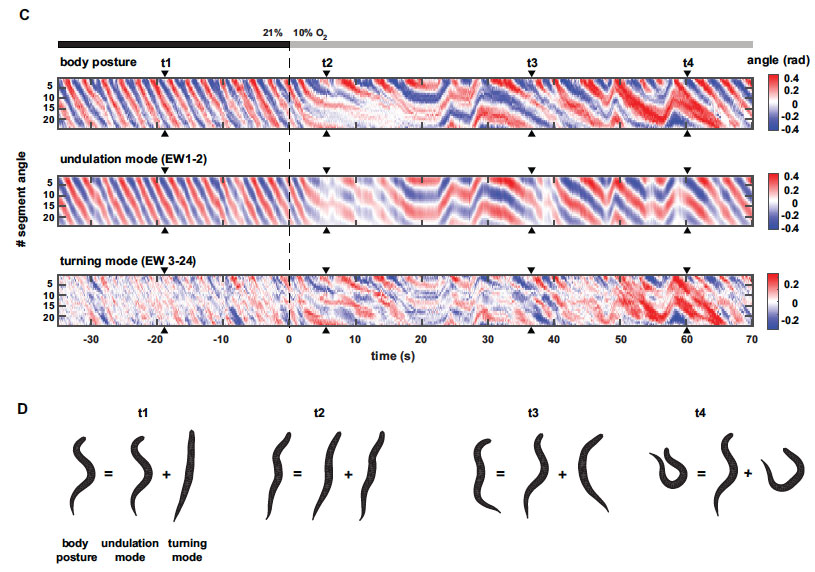 In animal locomotion a tradeoff exists between stereotypy and flexibility: fast long-distance travelling (LDT) requires coherent regular motions, while local sampling and area-restricted search (ARS) rely on flexible movements. We report here on a posture control system in C. elegans that coordinates these needs. Using quantitative posture analysis we explain worm locomotion as a composite of two modes: regular undulations versus flexible turning. Graded reciprocal regulation of both modes allows animals to flexibly adapt their locomotion strategy under sensory stimulation along a spectrum ranging from LDT to ARS. Using genetics and functional imaging of neural activity we characterize the counteracting interneurons AVK and DVA that utilize FLP-1 and NLP-12 neuropeptides to control both motor modes. Gradual regulation of behaviors via this system is required for spatial navigation during chemotaxis. This work shows how a nervous system controls simple elementary features of posture to generate complex movements for goal-directed locomotion strategies.
In animal locomotion a tradeoff exists between stereotypy and flexibility: fast long-distance travelling (LDT) requires coherent regular motions, while local sampling and area-restricted search (ARS) rely on flexible movements. We report here on a posture control system in C. elegans that coordinates these needs. Using quantitative posture analysis we explain worm locomotion as a composite of two modes: regular undulations versus flexible turning. Graded reciprocal regulation of both modes allows animals to flexibly adapt their locomotion strategy under sensory stimulation along a spectrum ranging from LDT to ARS. Using genetics and functional imaging of neural activity we characterize the counteracting interneurons AVK and DVA that utilize FLP-1 and NLP-12 neuropeptides to control both motor modes. Gradual regulation of behaviors via this system is required for spatial navigation during chemotaxis. This work shows how a nervous system controls simple elementary features of posture to generate complex movements for goal-directed locomotion strategies.
Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy.
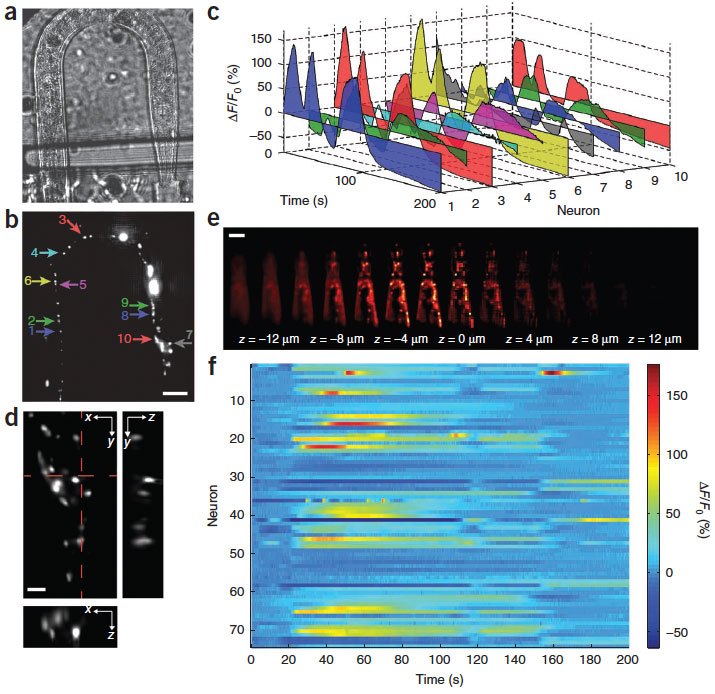 High-speed, large-scale three-dimensional (3D) imaging of neuronal activity poses a major challenge in neuroscience. Here we demonstrate simultaneous functional imaging of neuronal activity at single-neuron resolution in an entire Caenorhabditis elegans and in larval zebrafish brain. Our technique captures the dynamics of spiking neurons in volumes of ~700 mm x 700 mm x 200 mm at 20 Hz. Its simplicity makes it an attractive tool for high-speed volumetric calcium imaging.
High-speed, large-scale three-dimensional (3D) imaging of neuronal activity poses a major challenge in neuroscience. Here we demonstrate simultaneous functional imaging of neuronal activity at single-neuron resolution in an entire Caenorhabditis elegans and in larval zebrafish brain. Our technique captures the dynamics of spiking neurons in volumes of ~700 mm x 700 mm x 200 mm at 20 Hz. Its simplicity makes it an attractive tool for high-speed volumetric calcium imaging.
Temporal responses of C. elegans chemosensory neurons are preserved in behavioral dynamics.
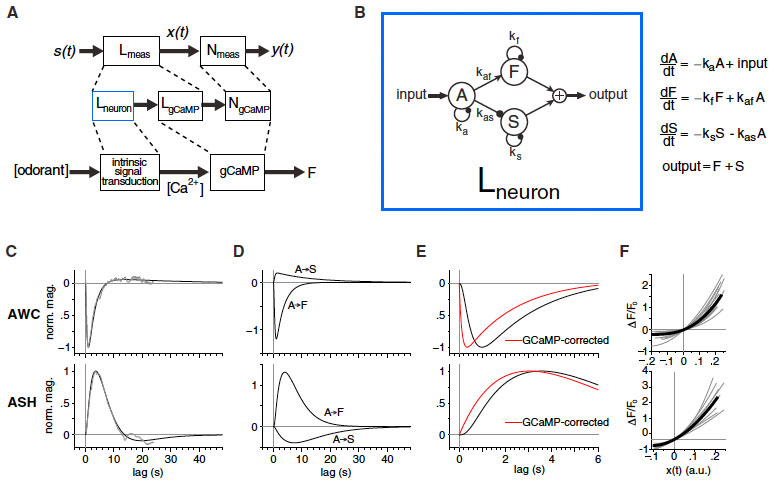 Animals track fluctuating stimuli over multiple timescales during natural olfactory behaviors. Here, we define mechanisms underlying these computations in Caenorhabditis elegans. By characterizing neuronal calcium responses to rapidly fluctuating odor sequences, we show that sensory neurons reliably
track stimulus fluctuations relevant to behavior.
AWC olfactory neurons respond to multiple odors
with subsecond precision required for chemotaxis,
whereas ASH nociceptive neurons integrate noxious
cues over several seconds to reach a threshold for
avoidance behavior. Each neuron's response to fluctuating
stimuli is largely linear and can be described
by a biphasic temporal filter and dynamical model. A
calcium channel mutation alters temporal filtering
and avoidance behaviors initiated by ASH on similar
timescales. A sensory G-alpha protein mutation affects
temporal filtering in AWC and alters steering
behavior in a way that supports an active sensing
model for chemotaxis. Thus, temporal features of
sensory neurons can be propagated across circuits
to specify behavioral dynamics.
Animals track fluctuating stimuli over multiple timescales during natural olfactory behaviors. Here, we define mechanisms underlying these computations in Caenorhabditis elegans. By characterizing neuronal calcium responses to rapidly fluctuating odor sequences, we show that sensory neurons reliably
track stimulus fluctuations relevant to behavior.
AWC olfactory neurons respond to multiple odors
with subsecond precision required for chemotaxis,
whereas ASH nociceptive neurons integrate noxious
cues over several seconds to reach a threshold for
avoidance behavior. Each neuron's response to fluctuating
stimuli is largely linear and can be described
by a biphasic temporal filter and dynamical model. A
calcium channel mutation alters temporal filtering
and avoidance behaviors initiated by ASH on similar
timescales. A sensory G-alpha protein mutation affects
temporal filtering in AWC and alters steering
behavior in a way that supports an active sensing
model for chemotaxis. Thus, temporal features of
sensory neurons can be propagated across circuits
to specify behavioral dynamics.

